
CDB2025 User Guide: Ortholog Mapping
CDB2025 User Guide: Ortholog Mapping
Purpose
Before exploring ligand–receptor interactions, you need to decide which species you want to use as your reference.
Ortholog mapping ensures that gene names in the database are matched correctly across species, so you can search and interpret results using the organism of interest.
This is especially important because although majority are, not all LR pairs in this database are originally human.
The database includes interactions from multiple species, and each table includes Evidence column:
- Direct – the LR pair has experimental support in the listed species.
- Inferred – the LR pair was inferred from an ortholog in another species.
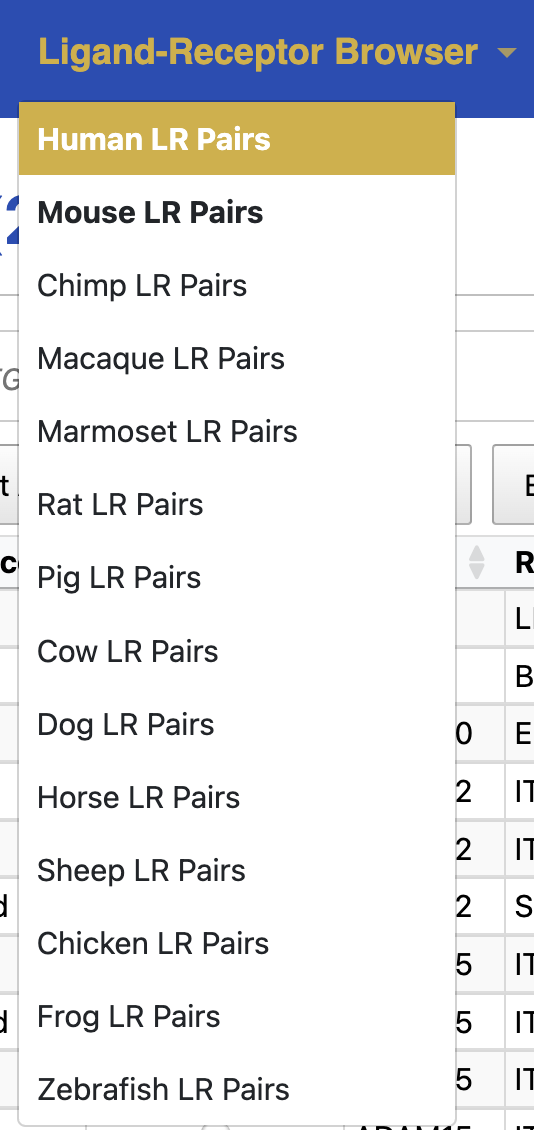
How to select your model organism
To select your model organism, follow these steps:
- Go to the Ligand-Receptor Browser on the navigation menu.
- Choose your target species from the drop-down menu.
- Available species include:
Human, Mouse, Chimpanzee, Macaque, Marmoset, Rat, Pig, Cow, Dog, Horse, Sheep (Rambouillet), Chicken, Frog, Zebrafish
- Available species include:
- Explore the Pair-list view table of your choice.
Once selected, The {Species} LR Pair will use gene names mapped to your chosen organism along with their annotations.
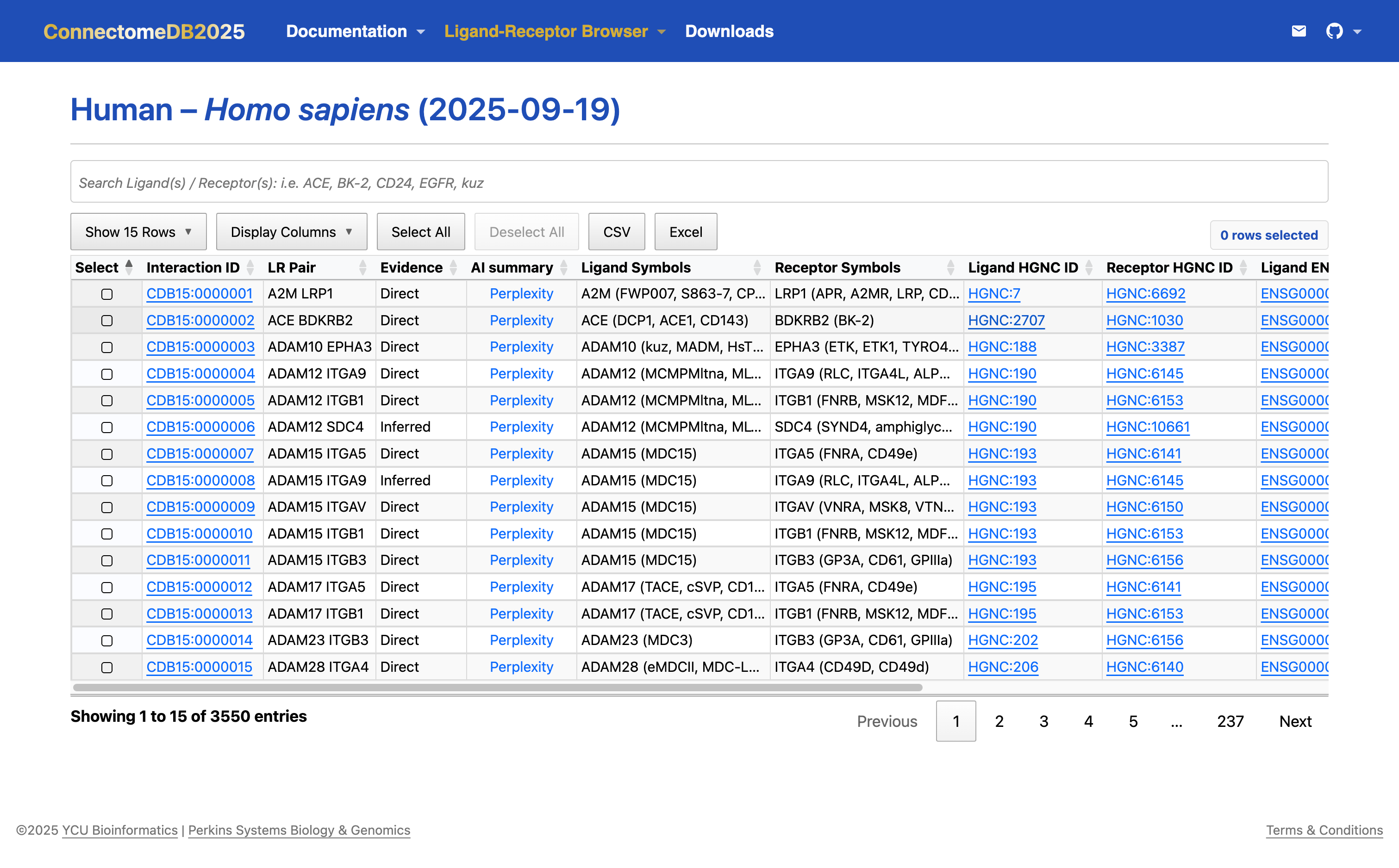
If you hover over the middle of each column header, you can see a tooltip description of the column.
If you hover over the middle of each cell within the Ligand symbols, and Receptor symbols columns, you can see the official gene symbol and its aliases and previous names within parentheses.
If you want to make more rows visible, adjust from the default 10 to 25, 50, or 100 rows per page using the Show 15 Rows drop-down menu.
If you want to make more columns visible, adjust using the Display Columns drop-down menu.
Some columns are links; Species Identifiers (i.e. HGNC, MGI, ZFIN, XEN, RGD, and XX ID) and AI summary links to external resources while Ligand-Receptor (LR) Pair Identifier (Interaction ID) links to the LR Pair Cards view.
See short clip showcasing the interactivity of the Pair-list view:
How the mapping works
- Ortholog relationships are sourced from Ensembl BioMart, NCBI, HGNC Comparison of Orthology Predictions (HCOP) and species-specific databases (HGNC, MGI, RGD, ZFIN, XEN). Additional species-specific databases may be incorporated in the future to further validate orthologs. For this reason, some columns in the species-specific tables—such as “Ligand XX ID” and “Receptor XX ID”—are currently empty and represented by “-”.
- The mapping preserves 1:1 orthologs where possible. If multiple orthologs exist, they are removed.
- Genes without orthologs are excluded from the results.
Tip
If you’re working exclusively in human datasets, you can leave the setting as Human. If you’re in doubt, choose the organism that matches your raw data source.
Previous: Overview Get an overview of the ConnectomeDB platform tutorial.
Next: Gene Search Look up genes in the pair list view.