CDB2025 User Guide: Download Data
CDB2025 User Guide: Download Data
Purpose
The ConnectomeDB provides a comprehensive collection of ligand–receptor interactions, which can be downloaded for offline analysis or integration into your own research workflows. This page guides you through the process of downloading data from the ConnectomeDB.
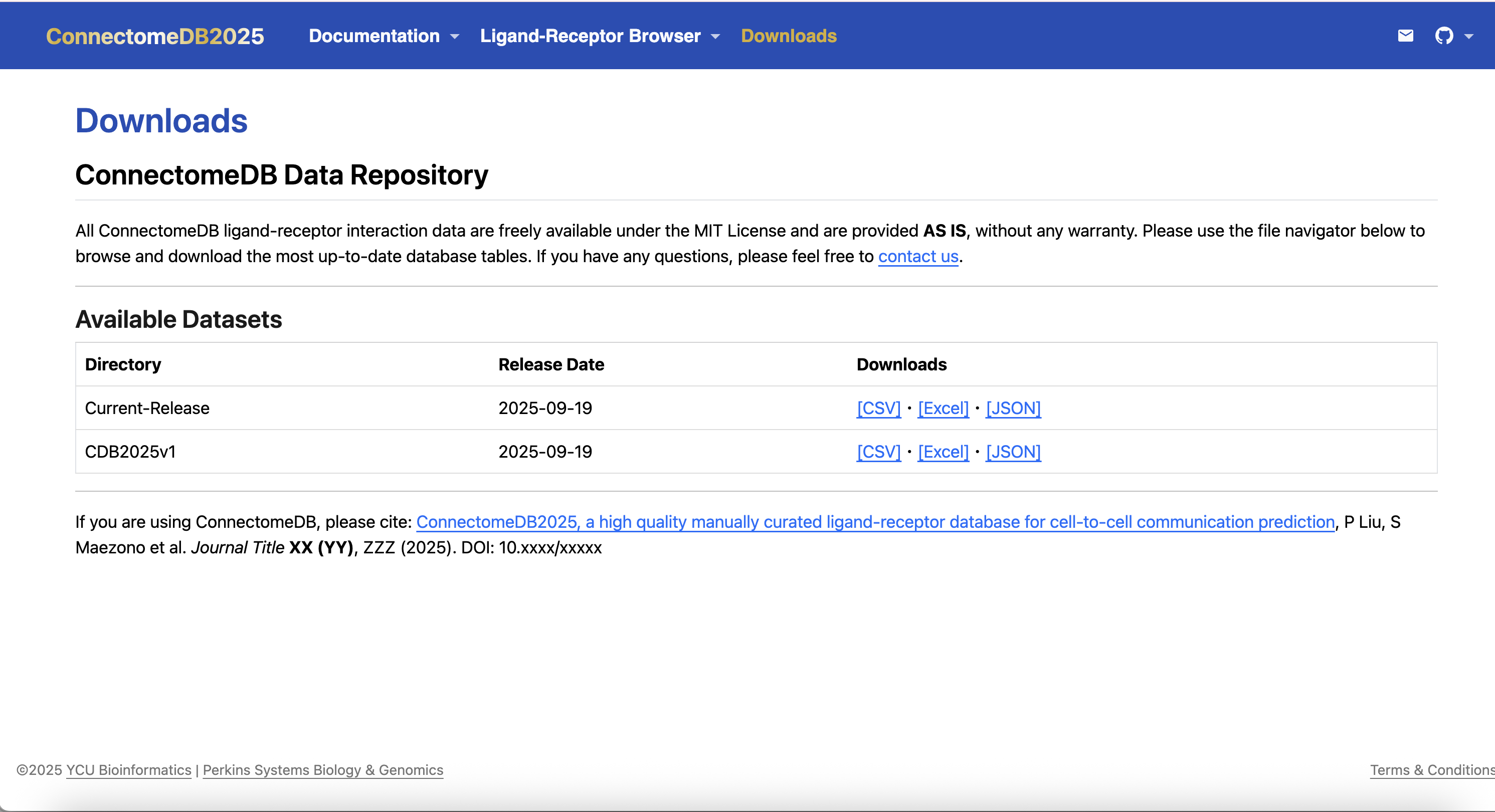
The Downloads provides access to bulk downloads of the ligand–receptor interaction data, organized by species and release version. It is designed to be user-friendly, allowing you to quickly find and access the data you need.
How to download data
To download data from the ConnectomeDB, follow these steps:
- Visit the Download Page: Go to the Downloads page on the ConnectomeDB website.
- Choose the Release Version: Select the specific release version of the data you wish to download. The ConnectomeDB provides versioned datasets, allowing you to access historical data or the latest updates as needed.
- Each release is date-stamped, ensuring reproducibility and clarity on the data’s temporal context.
- The latest release is labeled as “Current-Release” for easy identification.
- Select the Data Format: Choose the format you prefer for downloading the data. The ConnectomeDB offers data in various formats, including CSV, Excel (XLSX) and JSON, to accommodate different analysis needs.
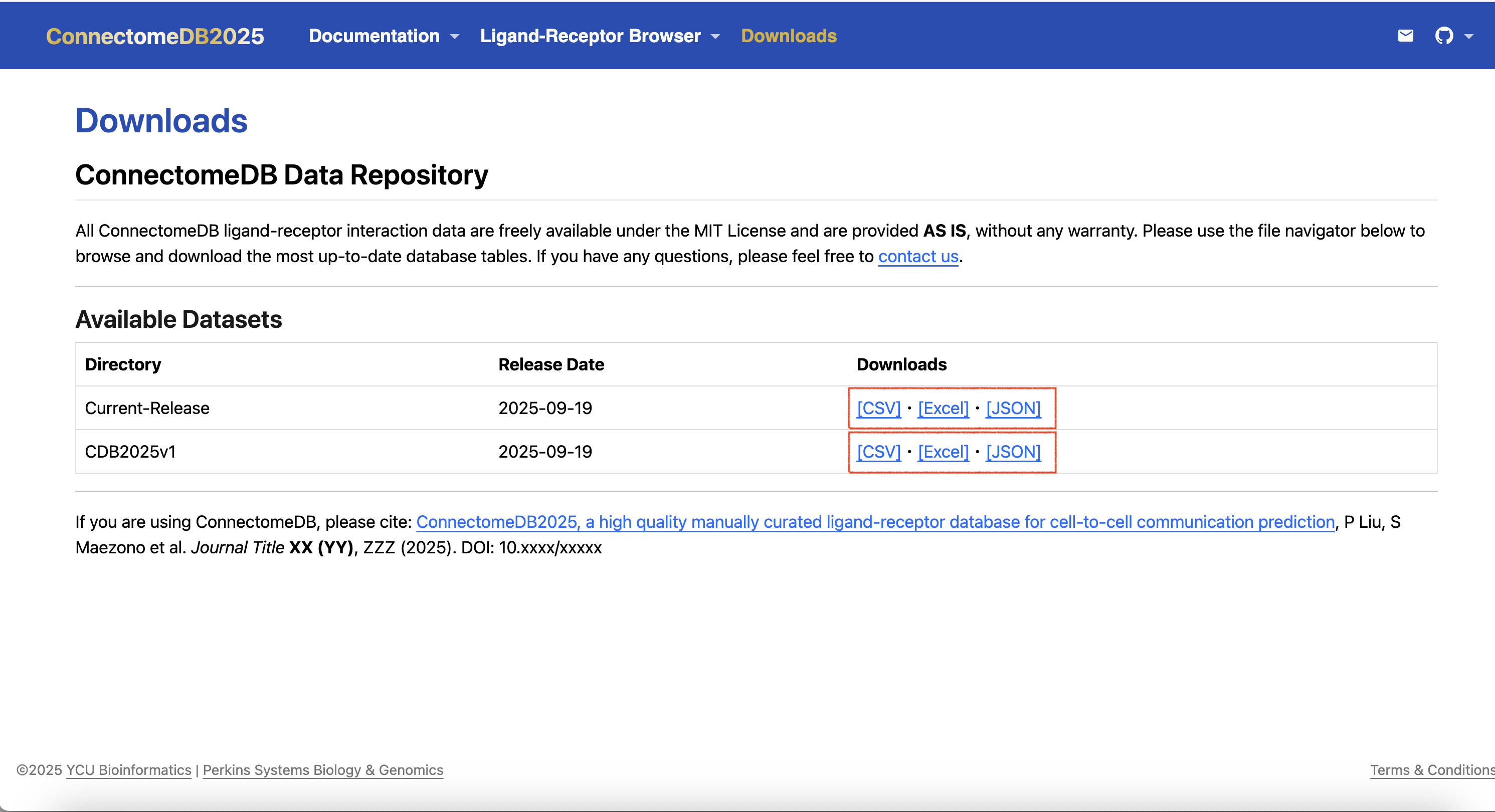
- Select the Species: Choose the species for which you want to download the ligand–receptor interaction data. The ConnectomeDB supports multiple species, including human and 13 additional model organisms.
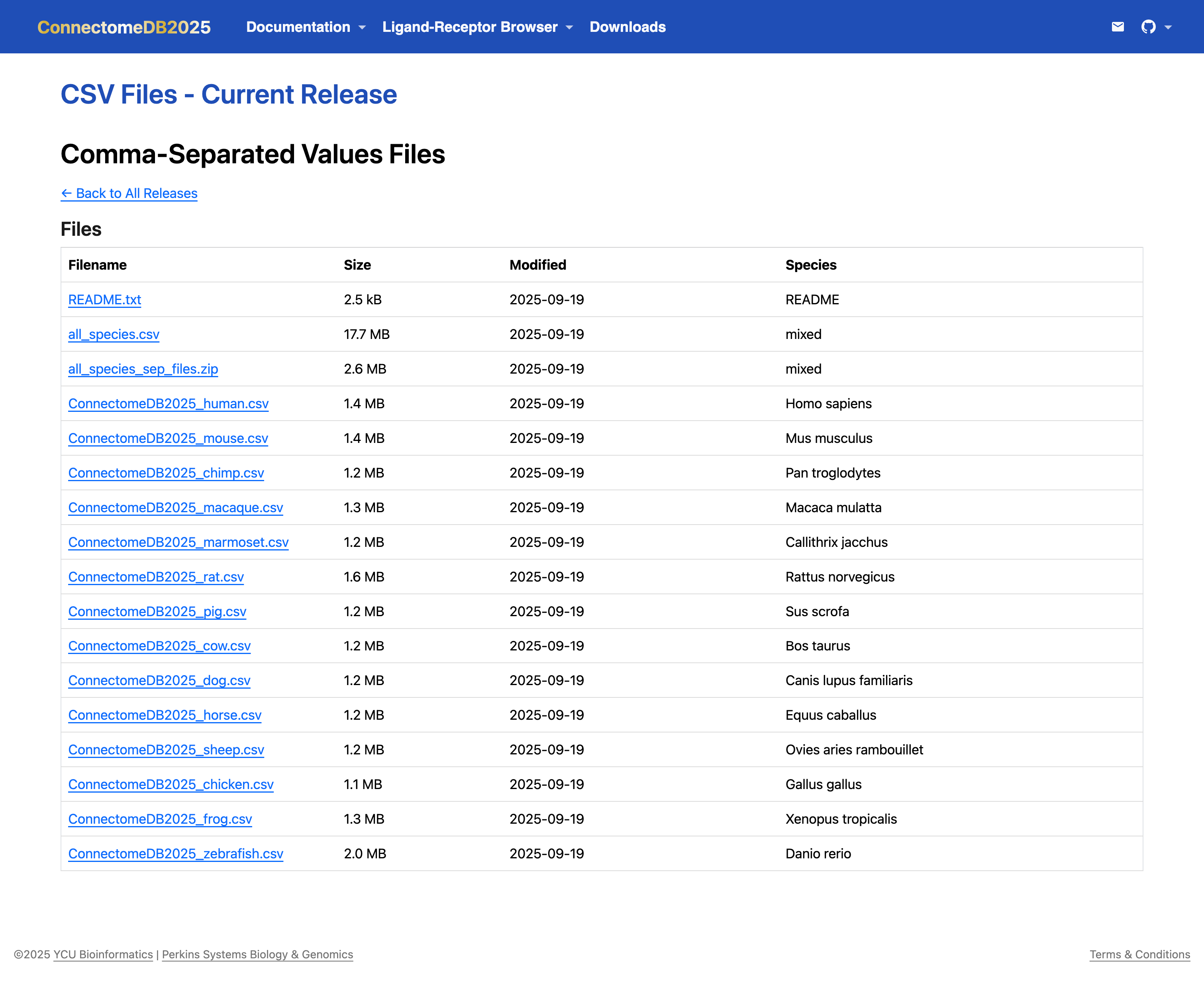
- Download the Data: Click the Filename to save the data file to your local machine. The file will contain all ligand–receptor interactions for the selected species and release version, along with relevant annotations.
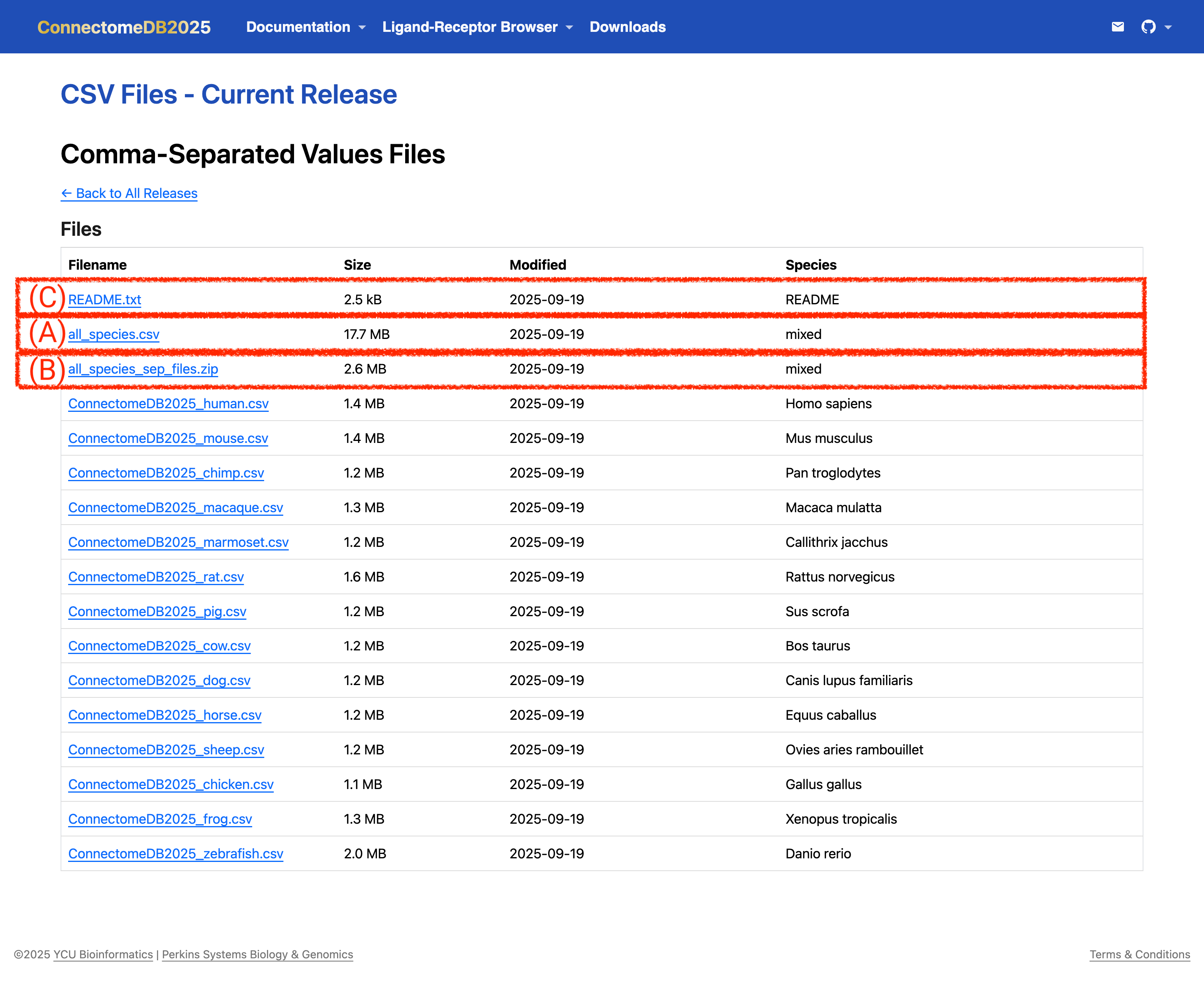
If you prefer one table/file that includes all species, you can download the
all_speciesfile in your preferred format (CSV, Excel(XLSX), or JSON). It contains 15 columns including aSpeciescolumn that identifies the species for each ligand–receptor pair. The individual IDs— Ligand {HGNC/MGI/RGD/XEN/ZFIN/XX} ID —are combined into the Ligand Species ID column, and Receptor {HGNC/MGI/RGD/XEN/ZFIN/XX} ID are combined into the Receptor Species ID column. See (A) in the image below.For bulk downloads, you can download a zipped folder,
all_species_sep_files.zip, which contains the data tables for all 14 species in your chosen format (CSV, Excel/XLSX, or JSON). Note that this does not include theall_speciesfile. (B) in the image below.
- Download the README: The
README.txtfile provides important information about the dataset, including column descriptions, data sources, and any relevant notes. It is recommended to download and review this file to better understand the structure and content of the data you are working with. See (C) in the image below.
How to download filtered data
If you want to download a subset of the data based on selected rows, see Gene Search for instructions on how to filter the data by specific genes or ligand–receptor pairs. Once you have applied your filters, you can download the filtered results directly from the Pair-list view.
Previous: LR Pair Card View Look up genes in the pair list view.
Next: User Submission Learn how to submit your own ligand–receptor pairs to the database.